Multivariate tensor-based ventricular morphometry analysis system
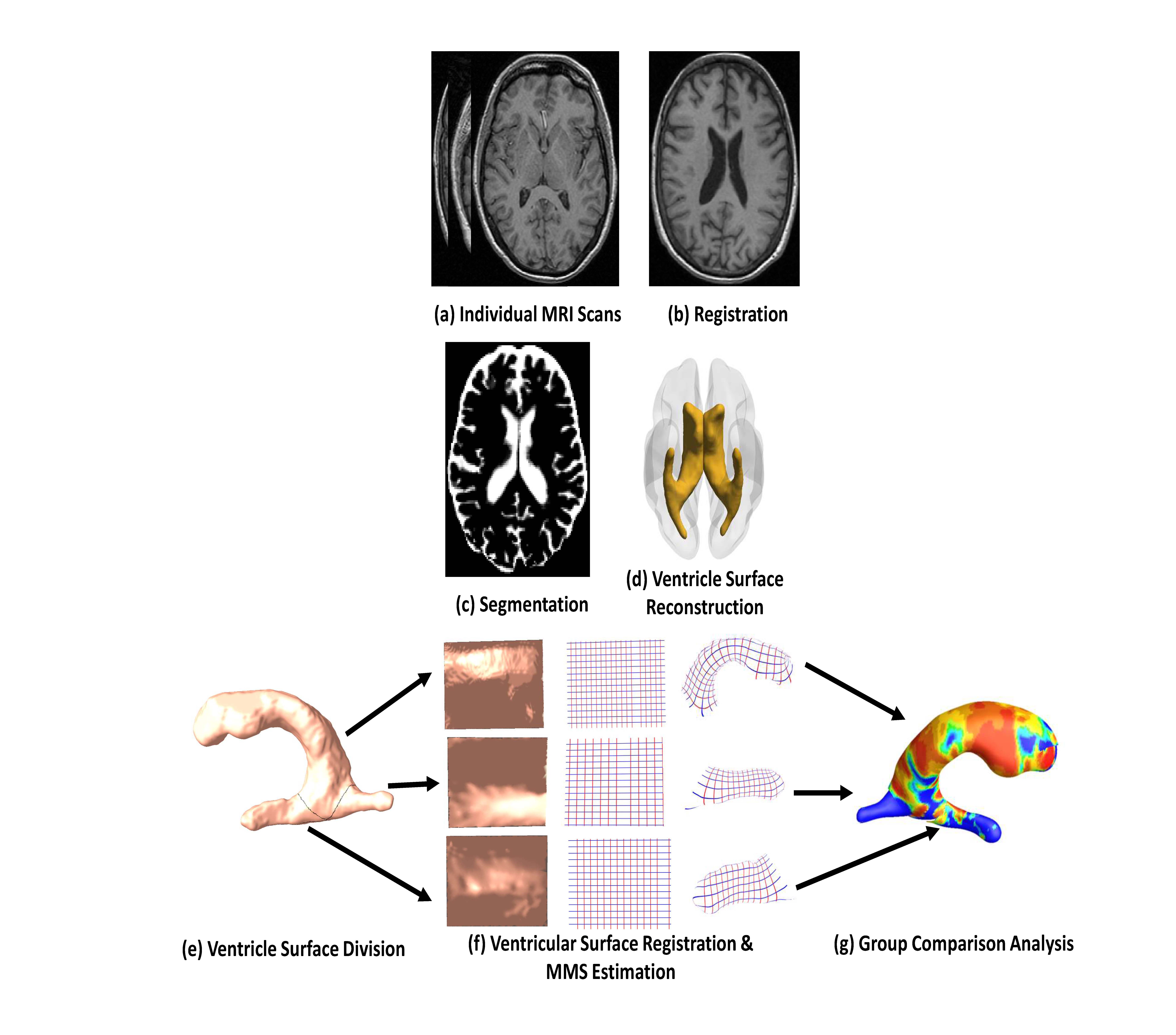
We introduce a ventricular morphometry analysis system (VMAS) that generates a whole connected 3D ventricular shape model and encodes a great deal of ventricular surface deformation information that is inaccessible by VV. VMAS contains an automated segmentation approach and surface-based multivariate morphometry statistics.
The whole pipeline is special for server mode. That means you can parallel deal with individual MRIs. The terminate results of “VMAS/run.py” are smoothed bilateral ventricles and can be used to do group-wise morphometry analysis as hippocampus pipeline (http://gsl.lab.asu.edu/software/mtbm-sma/), directional analysis and effect size analysis.
Additional software
Confirm your server has the Additional software:
- FSL/5.0.1
- Matlab/2017a
- Python 2.7
Set the parameters and paths for Ventricle data process
Please refer “run.py” to set the input path (including *.nii), output path (for storing ventricular results) and nodes (the server nodes for computing). The file ‘template.sh’ is an example of server setting information, run.py will send executable scripts to ‘template.sh’ for server computing.
Run ‘python run.py’ to process ventricles.
Note: Check the processing information in “<some directory>/VMAS/*.err and *.out”, the intermediate results can be observed in <some directory>/VMAS/tmp”. After getting the ventricles, you can remove these temporary results for saving space.
If you can not generate ventricles, you probably need to create the new template “Ave_Template_4.nii” using “tmp/<temporary folders>/rp3*nii”. The procedure: Matlab->load SPM path and run it->select “fMRI”->select “Batch”->spm->tools->shooting (create template) using all “rp3*.nii” files->create template->generate the new ‘Ave_Template_4.nii’->> replace ‘Ave_Template_4.nii’ under “<path>/VMAS/”
Group analysis
A similar command as:

Excel file stores the group information ‘grp0’ or ‘grp1’ of each subject like:

Directional and effect size analysis
Under ‘VMAS/tools’, there are Matlab scripts for directional analysis and effect size analysis.
Reference:
Dong Q, Zhang W, Stonnington CM, Wu J, Gutman BA, Chen K, Su Y, Baxter LC, Thompson PM, Reiman EM, Caselli RJ, Wang Y, ADNI Group, Applying Surface-Based Morphometry to Study Ventricular Abnormalities of Cognitively Unimpaired Subjects Prior to Clinically Significant Memory Decline, NeuroImage: Clinical, 2020, 27:102338