Univariate Neurodegeneration Biomarker Generation Framework
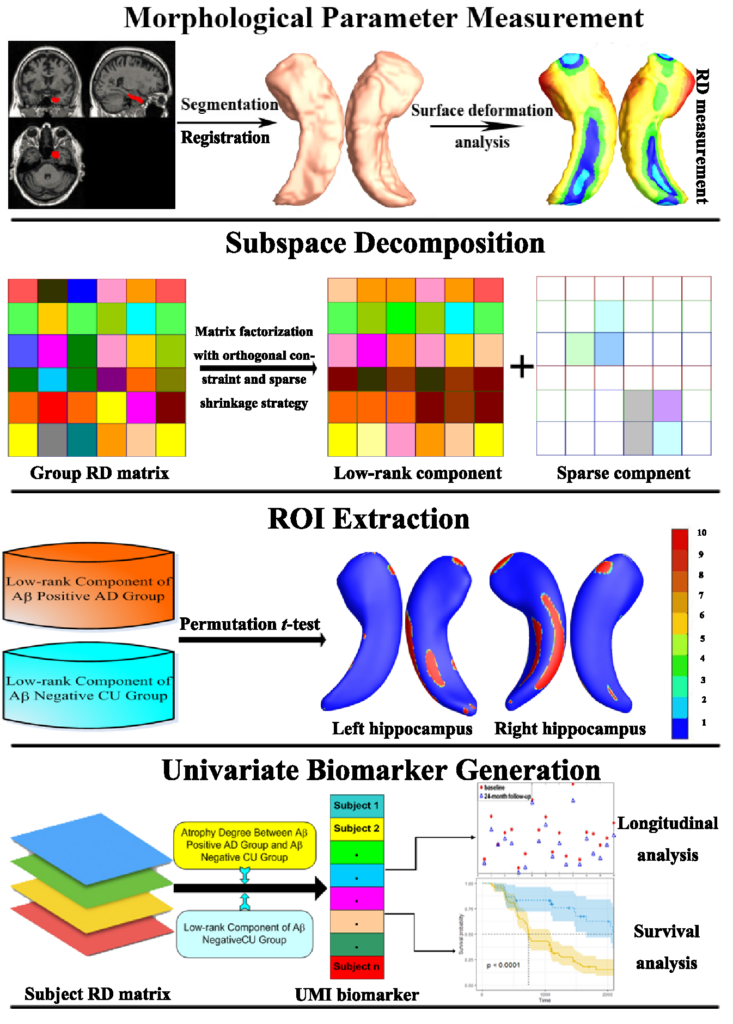
We develop a univariate neurodegeneration biomarker generation framework based on subspace decomposition to correctly and effectively depict the morphological changes induced by neurodegenerative diseases. Applying the matrix decomposition and the local sparse constraints on the radial distance (RD) observation matrices, we obtain the ROIs closely associated with neurodegnerative disease which are robust to image noise. We then generate the univariate morphometry index (UMI) to improve the statistical power for in vivo MRI morphological analyses.
1. Generate the statistical morphological measurement
Please use our software at http://gsl.lab.asu.edu/software/mtbm-sma/ to calculate the hippocampal surface features. Because hippocampal atrophy and enlargement directly affect the distance from each surface point to its medial core (analogous to the center line in a tube), we extracted this distance, i.e, the radial distances (RD), as the statistical morphological measurement. The RD extraction algorithm is also available at: http://gsl.lab.asu.edu/software/mtbm-sma/.
2. Extract the low-rank components
Please download the UMI package. When the RDs of the subjects belonging to the same group are stacked into columns to generate an observation matrix, the observation matrix is low-rank, and the low-rank components correspond to the group common morphological structure. According to subspace decomposition frame, the low-rank component L can be extracted based on the well-known alternating direction method and the Douglas-Rachford operator splitting(DR) method. The low-rank component extraction algorithm is available in UMI/low_rank_component_extraction.m. Scripts of the proposed algorithm is developed in MATLAB 2012 (or above version).
3. Generate ROIs
After extracting the low-rank components of Ab positive Alzheimer’s disease (AD) group and Ab negative cognitive unimpaired (CU) group, ROIs can be generated based on permutation t-test which is a type of statistical significance test. The distribution of the test statistic under the null hypothesis was obtained by calculating all possible values under rearrangements of the labels on the observed data points. The permutation t-test is available at: http://gsl.lab.asu.edu/software/mtbm-sma/.
4. Generate Univariate Univariate Morphometry Index (UMI)
When we obtain the voxel-wise AD atrophy weights by normalizing the mean differences between the L of CU group and the L of AD group on the predefined ROIs. The degree of atrophy can be computed in the predefined ROIs. Last, the UMIs can be obtained through the voxel-wise summation across all the voxels on the predefined ROIs with the production of AD atrophy weights and the atrophy degree of the testing individual. The UMIs computation algorithm is available at: UMI/generate_UMI.m. Scripts of the proposed algorithm is developed in MATLAB 2012 (or above version).