Brain Surface Parameterization using Riemann Surface Structure
Yalin Wang, Xianfeng Gu, Kiralee M. Hayashi, Tony F. Chan, Paul M. Thompson and Shing-Tung Yau
Abstract
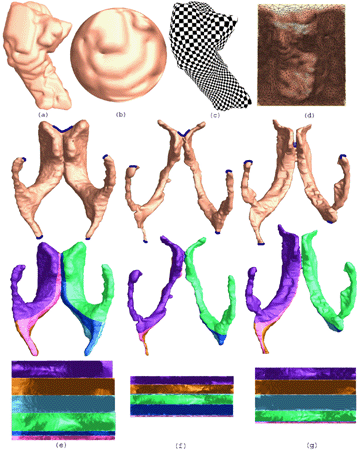
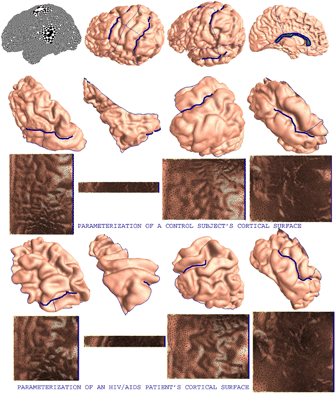
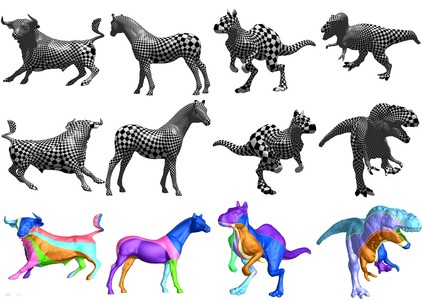
We develop a general approach that uses holomorphic 1-forms to parameterize anatomical surfaces with complex (possibly branching) topology. Rather than evolve the surface geometry to a plane or sphere, we instead use the fact that all orientable surfaces are Riemann surfaces and admit conformal structures, which induce special curvilinear coordinate systems on the surfaces. Based on Riemann surface structure, we can then canonically partition the surface into patches. Each of these patches can be conformally mapped to a parallelogram. The resulting surface subdivision and the parameterizations of the components are intrinsic and stable. To illustrate the technique, we computed conformal structures for several types of anatomical surfaces in MRI scans of the brain, including the cortex, hippocampus, and lateral ventricles. We found that the resulting parameterizations were consistent across subjects, even for branching structures such as the ventricles, which are otherwise difficult to parameterize. Compared with other variational approaches based on surface inflation, our technique works on surfaces with arbitrary complexity while guaranteeing minimal distortion in the parameterization. It also offers a way to explicitly match landmark curves in anatomical surfaces such as the cortex, providing a surface-based framework to compare anatomy statistically and to generate grids on surfaces for PDE-based signal processing.
Figures (click on each for a larger version):
Related Publications
- Y. Wang, L.M. Lui, X. Gu, K.M. Hayashi, T.F. Chan, A.W. Toga, P.M. Thompson and S.-T. Yau, Brain Surface Conformal Parameterization Using Riemann Surface Structure“, IEEE Transactions on Medical Imaging, Vol. 26, Nov. 6, June 2007 pp. 853-865
- Y. Wang, X. Gu, K.M. Hayashi, T.F. Chan, P.M. Thompson and S.-T. Yau, “Surface Parameterization using Riemann Surface Structure“, 10th IEEE International Conference on Computer Vision (ICCV), Beijing, China, Oct. 2005, pp. 1061-1066